Courtney Davis Contributes to Computational SARS-CoV-2 Tissue Model
 Seaver College associate professor of mathematics Courtney Davis joined a multi-disciplinary
group of scientists to build, expand and test a computational model that simulates
the viral replication and within-host spread of SARS-CoV-2, the novel virus that causes
COVID-19 disease.
Seaver College associate professor of mathematics Courtney Davis joined a multi-disciplinary
group of scientists to build, expand and test a computational model that simulates
the viral replication and within-host spread of SARS-CoV-2, the novel virus that causes
COVID-19 disease.
Built by Paul Macklin from Indiana University’s Luddy School of Informatics, Computing, and Engineering, the PhysiCell framework was originally designed to study complex systems problems like cancer growth within tissues. On March 25, Macklin worked through the night adapting the system to simulate SARS-CoV-2. After he live-tweeted his progress, Davis and scientists around the world began providing feedback and offering support. In a matter of days, a group of scientists united to contribute to Macklin’s rapid-prototyping project and drive it towards a sustainable, community-driven effort.
“It’s empowering to see scientists coming together to collectively solve these problems,” shares Davis. “This project wouldn’t have been possible without Macklin’s collaborative approach and eagerness to keep it open-sourced and to seek feedback from others.”
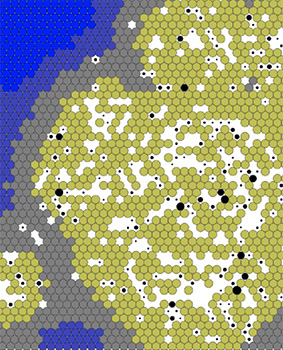
The multiscale model tracks the virus in a close-up, cellular view of a sheet of epithelial lung tissue. It shows the behaviors of the virions as they replicate within a cell and disperse to neighboring cells. The interactive model aims to provide valuable insights not only into the dynamics of the virus but also into the potential interactions of the virus with treatments and drug combinations. While the model is still preliminary and is not currently used for clinical decisions, it is an important stride to increase scientific understanding of the behavior of SARS-CoV-2 within tissue cells.
The group’s expeditious work allowed for a second version of the model to be released shortly after the first model. The update now allows for modular functionality so groups may build and test different parts of the model collaboratively and in parallel. The model will remain open-source, and, as the group continues to test and validate the model against biological data, they aim to release new versions with increased functionality in a 2-3 week cycle.
To learn more about the group’s collaborative work, read the project’s preprint and explore the interactive model.